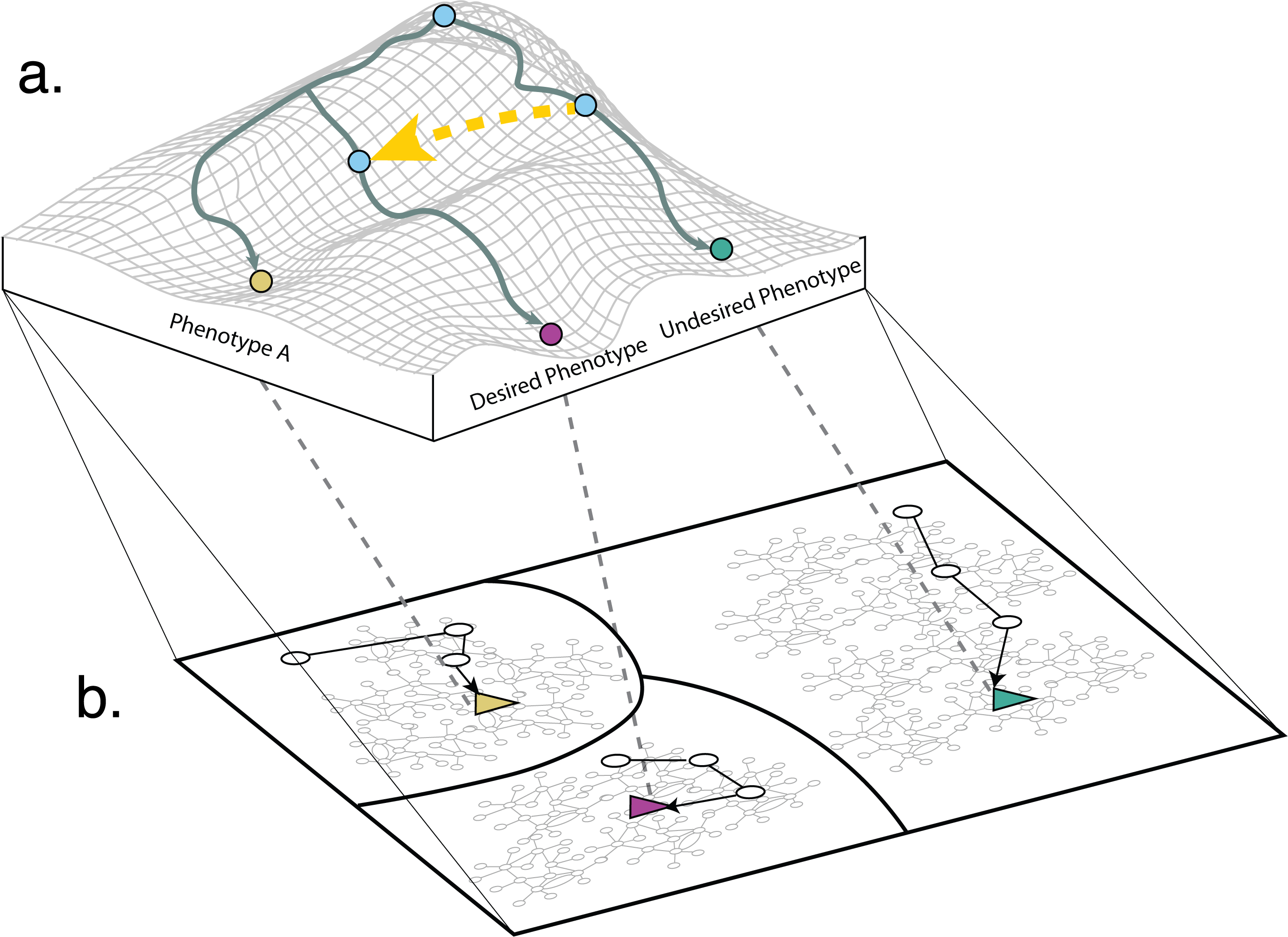
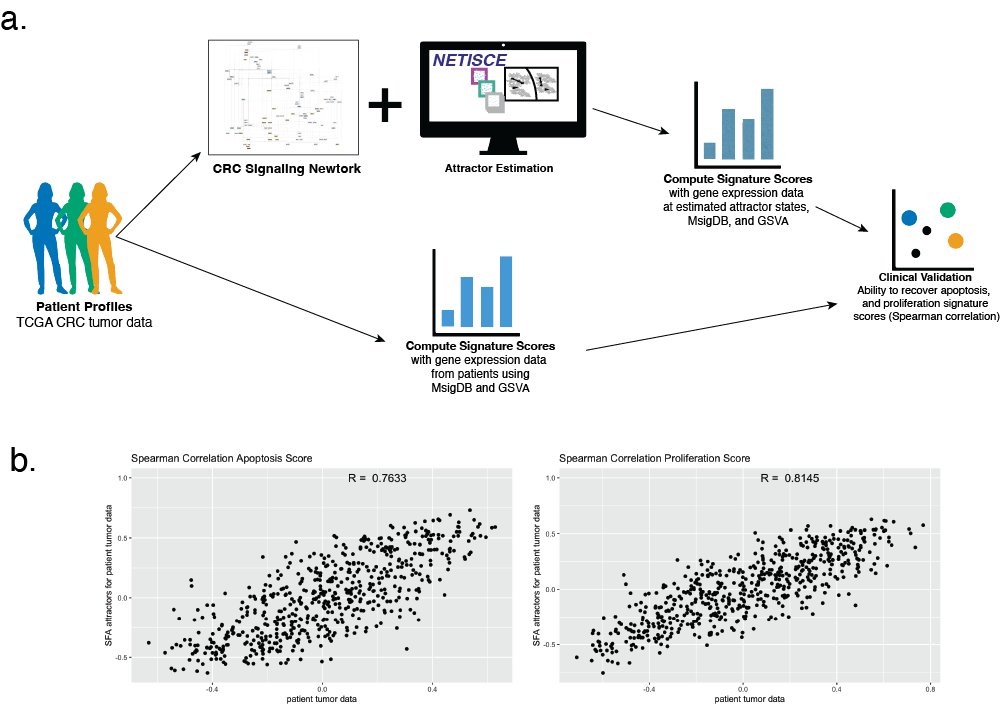
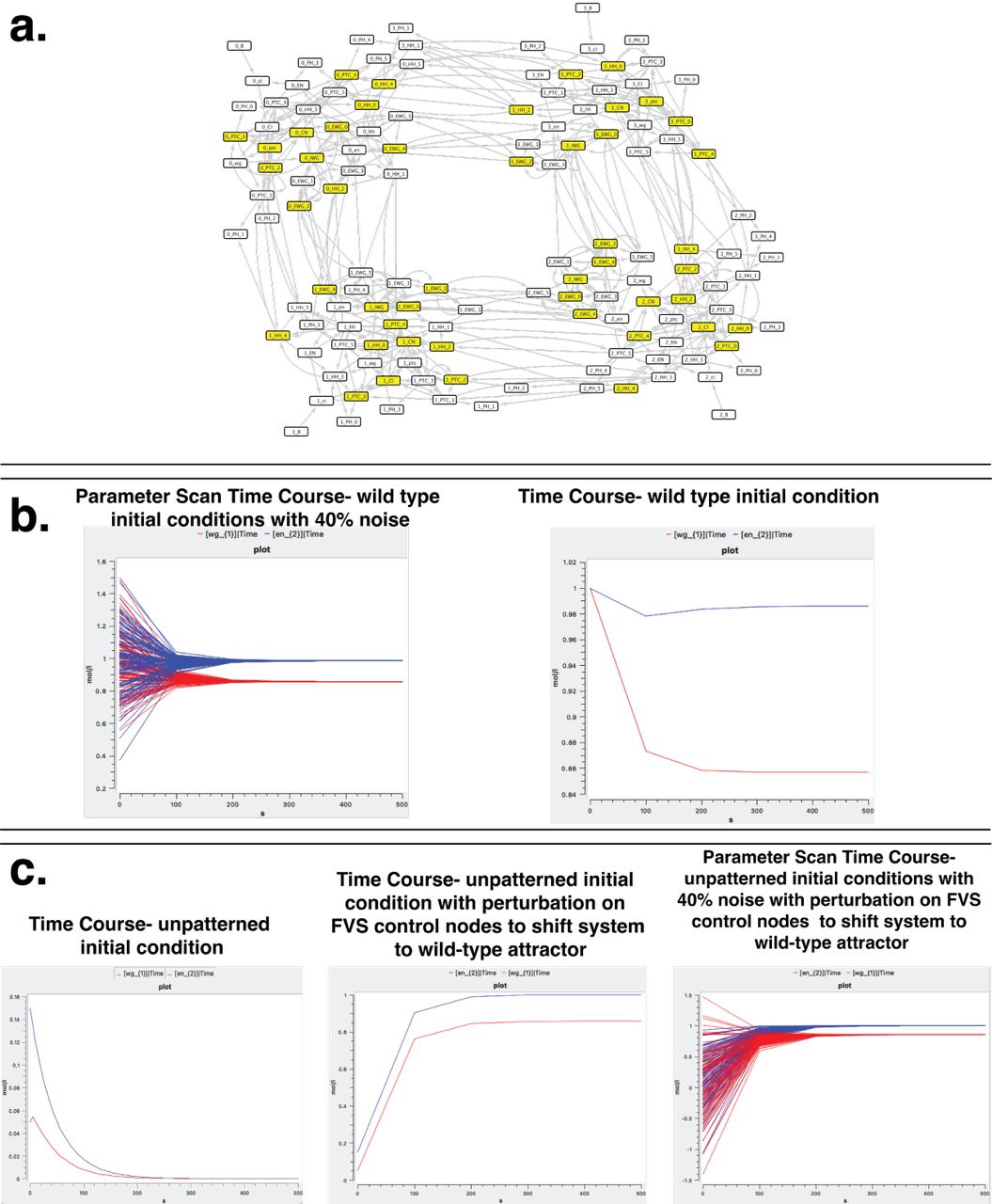
Welcome to the Vera-Licona Lab
At the intersection of computational systems biology, systems medicine, mathematical biology, and bioinformatics, our research lab is dedicated to developing and applying mathematical algorithms for both modeling and reprogramming of biological systems. Central to our work is the integration of multiomics data, emphasizing data-driven methodologies.
Past and current collaborative application areas include cancer research, stem cell research, immunology and biogerontology (see Research).
We are located at the Center for Quantitative Medicine, which is part of the Center for Cell Analysis & Modeling.
We are always looking for passionate students at diverse levels and Postdocs to join the team(more info)!
News
March 19, 2024
Dr. Vera-Licona will give an invited talk at the Math Department, University of California, Riverside on May 1st, 2024
January 3, 2024
Dr. Vera-Licona will give an invited talk in the special session Computational Biomedicine: Methods – Models – Applications at the Joint Mathematics Meetings (JMM) 2024 in San Francisco, CA.
November 8, 2023
Dr. Vera-Licona will give an invited talk at the Inauguration of the Mexican Biology and Mathematics Network (RedMexBiomate) in Cuernavaca, Mexico
October 11, 2023
Dr. Vera-Licona will present their efforts to construct an Encyclopedia of Cancer Cell Reversion at the International Conference of Systems Biology (ICSB 2023) within the Systems Biology of Cancer Session
October 11, 2023
Today, Lauren Marazzi will introduce NETISCE at the International Conference of Systems Biology (ICSB 2023) within the Network Biology Session
March 20, 2023
Dr. Vera-Licona will give an invited talk in June at the Centre International de Recontres Mathématiques (CIRM) in Marseille, France during the workshop on Networks and Biological Models Inference in the CNRS Bioss working group on symbolic systems biology
October 13, 2022
Dr. Vera-Licona will give a talk today at the International Conference of Systems Biology (ICSB 2022) in Berlin within the Machine Learning & Artificial Intelligence Session